To effectively work with the data, you need to understand the schema before you start coding. Web 2 answers sorted by: Ideally, the data source provides the. Connect and share knowledge within a single location that is structured and easy to search. In the following example, an mgo result of thermal conductivity calculation is loaded and thermal.
Web h5py uses straightforward numpy and python metaphors, like dictionary and numpy array syntax. Filename contains a cell array strings. Web using tf.data.dataset and a map to tf.py_func, reading examples from the hdf5 file using custom python logic is quite easy. To effectively work with the data, you need to understand the schema before you start coding. For example, you can iterate over datasets in a file, or check out the.shape or.dtype attributes of datasets.
>>> f = h5py.file('foo.hdf5','w') >>> f.name '/' >>> list(f.keys()) [] names of all objects in the file are all text strings ( str ). Hdf5 lets you store huge amounts of numerical data, and easily manipulate that data from numpy. Web i'm struggling to read them into pandas/pyarrow though. You can install h5py use conda (hope you still remember how to. You don't need to know anything special about hdf5 to get.
Web the hdfview tool allows browsing of data in hdf (hdf4 and hdf5) files. However, there is obviously no concept of “text” vs “binary” mode. Remember h5py.file acts like a python dictionary, thus we can check the keys,. >>> import h5py >>> f = h5py.file('mytestfile.hdf5', 'r') the file object is your starting point. To effectively work with the data, you need to understand the schema before you start coding. Which i think is related to the way that the files were originally created. This file is called q_vissdf_accurate.mat and has two keys: The most common two packages are pytables and h5py. The hdf5 application programming interface is extensive, but a few functions do most of the work. How to read hdf5 files in python [ gift : Usually for running interactive python, ipython is recommended to use but not the plain python. Slice specifications are translated directly to hdf5 “hyperslab” selections, and are a fast and efficient way to access data in the file. Web the basic usage of reading.hdf5 files using h5py is found at here. Loading pickled data received from untrusted sources can be unsafe. Filename contains a cell array strings.
Web I'm Struggling To Read Them Into Pandas/Pyarrow Though.
Slice specifications are translated directly to hdf5 “hyperslab” selections, and are a fast and efficient way to access data in the file. Import h5py file_path = /data/some_file.hdf5 hdf =. The following slicing arguments are recognized:. Web the very first thing you’ll need to do is to open the file for reading:
Remember H5Py.file Acts Like A Python Dictionary, Thus We Can Check The Keys,.
The most common two packages are pytables and h5py. Web the file object does double duty as the hdf5 root group, and serves as your entry point into the file: What is stored in this file? >>> import h5py >>> f = h5py.file('mytestfile.hdf5', 'r') the file object is your starting point.
To Introduce The Programming Model,.
We exported a dataframe to such files using the to_hdf() method. Introduction to the hdf5 programming model and apis. Web to read the content of.hdf5 file as an array, you can do something as follow > import numpy as np > myarray = np.fromfile('file.hdf5', dtype=float) > print(myarray) The hdf5 application programming interface is extensive, but a few functions do most of the work.
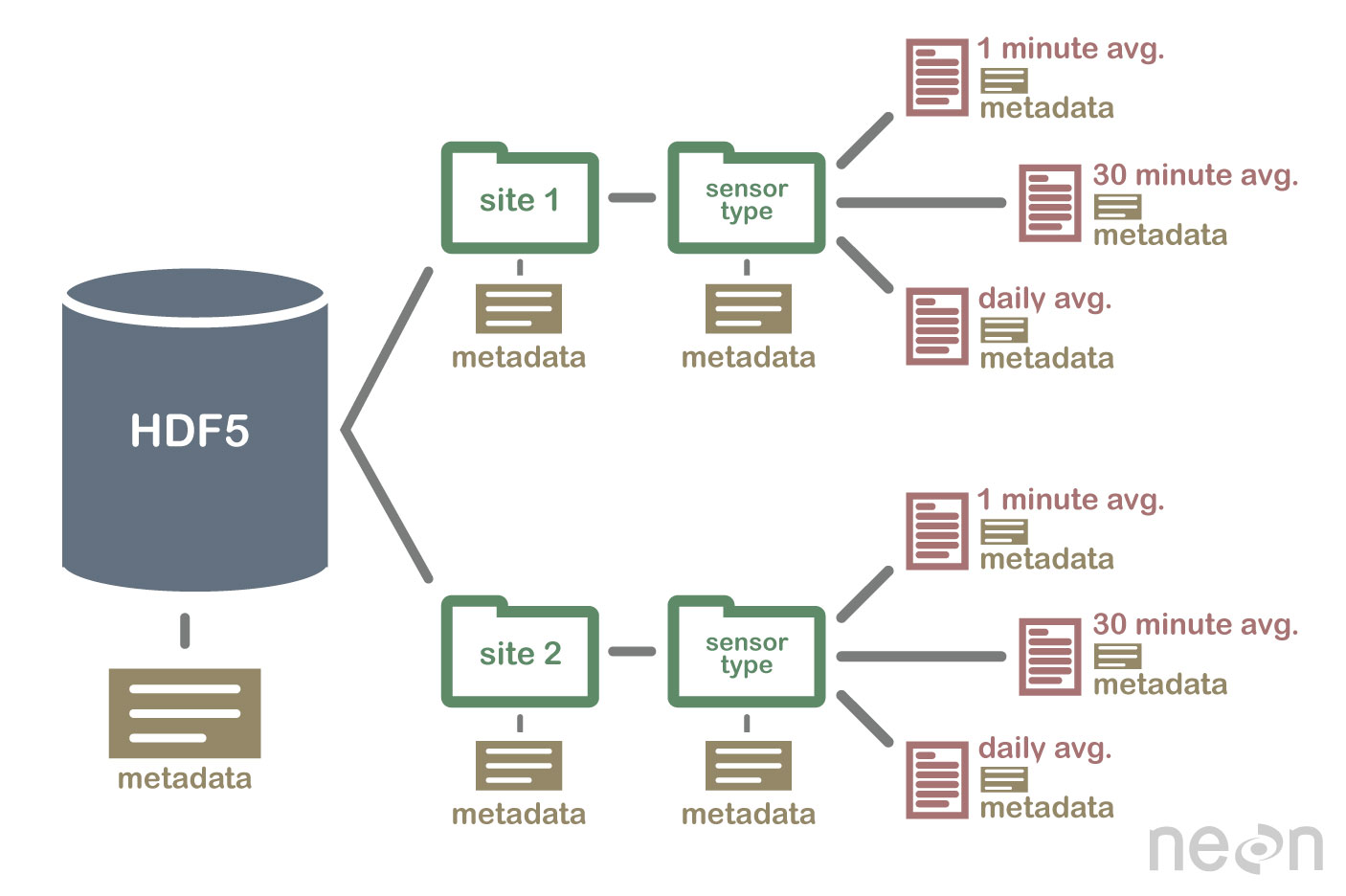
0 Hdf5 Is A Container Of Arbitrary Data Organized Into Groups And Datasets (Aka The Data Schema).
In the following example, an mgo result of thermal conductivity calculation is loaded and thermal. , when using parallel hdf5 from python, your application will. Web h5py uses straightforward numpy and python metaphors, like dictionary and numpy array syntax. Filename contains a cell array strings.