Most prokaryotic genes), an orf in the dna sequence will define the entire translated region. Web an open reading frame (orf) is a series of codons that begins with a start codon (usually aug) and ends with a stop codon. Web an open reading frame is a portion of a dna molecule that, when translated into amino acids, contains no stop codons. What it refers to is a frame of reference, and what is being read, reading, is the rna code, and it is being read by the ribosomes in order to make a protein. Where these triplets equate to amino acids or stop signals during translation, they are called codons.
Web every region of dna has six possible reading frames, three in each direction. Usually, this is considered within a studied region of a prokaryotic dna sequence, where only one of the six possible reading frames will be open (the reading, however, refers to the rna produced by transcription of the. Since each coding unit (codon) of the genetic code consists of three consecutive bases, the reading frame is established according to precisely where translation starts. Typically only one reading frame is used in translating a gene (in eukaryotes), and this is often the longest open reading frame. Web an open reading frame (orf) is a series of codons that begins with a start codon (usually aug) and ends with a stop codon.
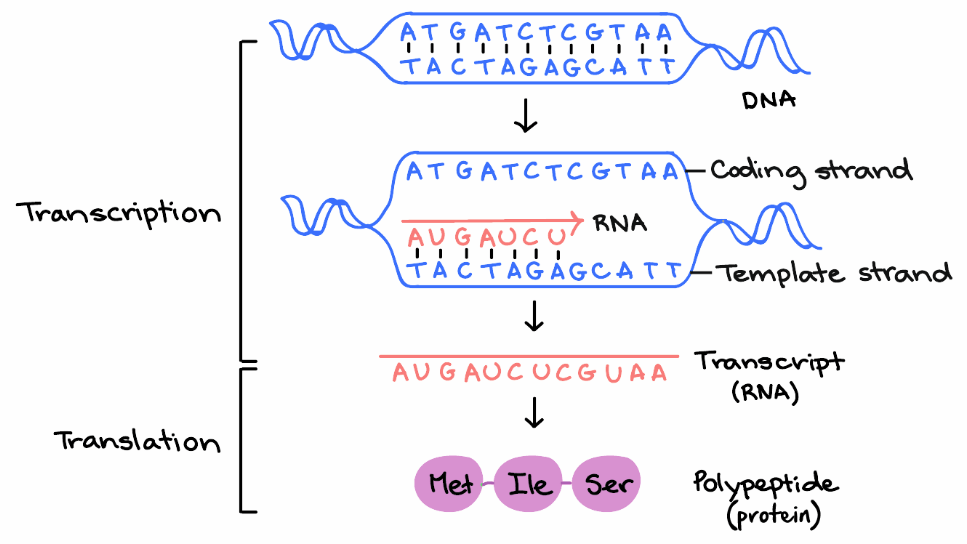
Where these triplets equate to amino acids or stop signals during translation, they are called codons. Web orf finder searches for open reading frames (orfs) in the dna sequence you enter. In genes that lack introns (e.g. That's a pretty abstract concept, so let's look at an example to understand it better. Web reading frame determines how the mrna sequence is divided up into codons during translation.
Most prokaryotic genes), an orf in the dna sequence will define the entire translated region. Web every region of dna has six possible reading frames, three in each direction. Web overview reading frame quick reference a sequence of bases in messenger rna (or deduced from dna) that encodes for a polypeptide. There can be no additional stop codons within that sequence. The genetic code reads dna sequences in groups of three base pairs,. A sequence of nucleotide triplets that is potentially translatable into a polypeptide and that is determined by the placement of a codon that initiates translation examples of reading frame in a sentence In genes that lack introns (e.g. Use orf finder to search newly sequenced dna for potential protein encoding segments, verify predicted protein using newly developed smart blast or regular blastp. The reading frame that is used determines which amino acids will be encoded by a gene. The mrna below can encode three totally different proteins, depending on the frame in which it's read: Typically only one reading frame is used in translating a gene (in eukaryotes), and this is often the longest open reading frame. Web an open reading frame is a portion of a dna molecule that, when translated into amino acids, contains no stop codons. Web in molecular biology, open reading frames ( orfs) are defined as spans of dna sequence between the start and stop codons. Since each coding unit (codon) of the genetic code consists of three consecutive bases, the reading frame is established according to precisely where translation starts. The program returns the range of each orf, along with its protein translation.
Web In Molecular Biology, Open Reading Frames ( Orfs) Are Defined As Spans Of Dna Sequence Between The Start And Stop Codons.
Web every region of dna has six possible reading frames, three in each direction. There can be no additional stop codons within that sequence. The program returns the range of each orf, along with its protein translation. Usually, this is considered within a studied region of a prokaryotic dna sequence, where only one of the six possible reading frames will be open (the reading, however, refers to the rna produced by transcription of the.
Web Overview Reading Frame Quick Reference A Sequence Of Bases In Messenger Rna (Or Deduced From Dna) That Encodes For A Polypeptide.
Web reading frame determines how the mrna sequence is divided up into codons during translation. The genetic code reads dna sequences in groups of three base pairs,. The mrna below can encode three totally different proteins, depending on the frame in which it's read: What it refers to is a frame of reference, and what is being read, reading, is the rna code, and it is being read by the ribosomes in order to make a protein.
Open Reading Frame Is A Terrible Term That We're Stuck With.
The reading frame that is used determines which amino acids will be encoded by a gene. Since each coding unit (codon) of the genetic code consists of three consecutive bases, the reading frame is established according to precisely where translation starts. Web an open reading frame is a portion of a dna molecule that, when translated into amino acids, contains no stop codons. Web orf finder searches for open reading frames (orfs) in the dna sequence you enter.
In Genes That Lack Introns (E.g.
Use orf finder to search newly sequenced dna for potential protein encoding segments, verify predicted protein using newly developed smart blast or regular blastp. Web an open reading frame (orf) is a series of codons that begins with a start codon (usually aug) and ends with a stop codon. Typically only one reading frame is used in translating a gene (in eukaryotes), and this is often the longest open reading frame. A sequence of nucleotide triplets that is potentially translatable into a polypeptide and that is determined by the placement of a codon that initiates translation examples of reading frame in a sentence