But they can also use orfs for more experimental purposes. Web how is open reading frame (orf) chosen? Web open reading frame (orf) is a basic term in molecular genetics and bioinformatics. The orf finder is a program available at ncbi website. Start codons are highlighted in purple, and stop codons are highlighted in red.
Web an open reading frame starts with an atg (met) in most species and ends with a stop codon (taa, tag or tga). Gene finding in organism specially prokaryotes starts form searching for an open reading frames (orf). Web 1 answer sorted by: Of these, only a fraction make their way into transcripts and only some of these end up being translated. Web how is open reading frame (orf) chosen?
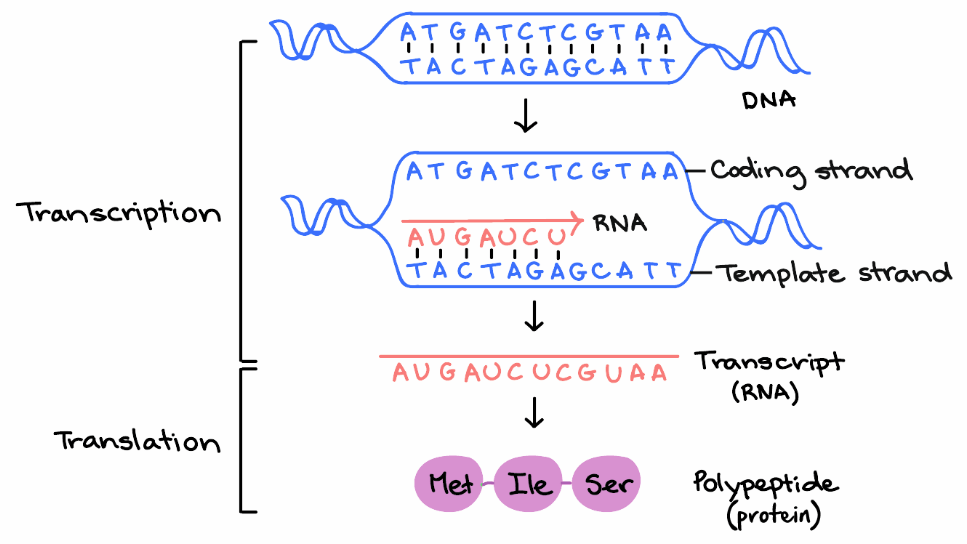
What it refers to is a frame of reference, and what is being read, reading, is the rna code, and it is being read by the ribosomes in order to make a protein. Orfs occur randomly and abundantly across the whole genome. In molecular biology, open reading frames ( orfs) are defined as spans of dna sequence between the start and stop codons. Web the region of a nucleotide that starts from an initiation codon and ends with a stop codon is called an open reading frame (orf). For example, the following sequence of dna can be read in six reading frames.
However, in some genes such as yeast gcn4, translation of specific uorfs may. Web orf finder searches for open reading frames (orfs) in the dna sequence you enter. Web an open reading frame starts with an atg (met) in most species and ends with a stop codon (taa, tag or tga). Web open reading frames (orfs) are parts of a reading frame that contain no stop codons. The three reading frames in the forward direction are shown with the translated amino acids below each. Web the term open reading frame (orf) is of central importance to gene finding. Of these, only a fraction make their way into transcripts and only some of these end up being translated. The program returns the range of each orf, along with its protein translation. Researchers can scan the genome for open reading frames to find genes that encode proteins. Web open reading frame (orf) is a basic term in molecular genetics and bioinformatics. An orf is a sequence of dna that starts with start codon “atg” (not always) and ends with any of the three termination codons (taa, tag, tga). The core protein (hbcag) is a product of 185 amino acids, encoded from the second start. But they can also use orfs for more experimental purposes. 2 i (mostly) like the definitions given on this page. Use orf finder to search newly sequenced dna for potential protein encoding segments.
The Orf Finder Is A Program Available At Ncbi Website.
Of these, only a fraction make their way into transcripts and only some of these end up being translated. Web an open reading frame starts with an atg (met) in most species and ends with a stop codon (taa, tag or tga). Proteins are formed from orf. Orfs occur randomly and abundantly across the whole genome.
Uorfs Can Regulate Eukaryotic Gene Expression.
An orf is a sequence of dna that starts with start codon “atg” (not always) and ends with any of the three termination codons (taa, tag, tga). Web the region of a nucleotide that starts from an initiation codon and ends with a stop codon is called an open reading frame (orf). 2008 ), built by imgt®, the international. Surprisingly, at least three definitions are in use.
2 I (Mostly) Like The Definitions Given On This Page.
The region of the nucleotide sequences from the start codon (atg) to the stop codon is called the open reading frame. Web open reading frame (orf) 1 of the px region encodes p12 (fig. Eukaryotic messenger rnas predominantly have a single main. Open reading frame is a terrible term that we're stuck with.
The Three Reading Frames In The Forward Direction Are Shown With The Translated Amino Acids Below Each.
Web two open reading frames (orfs) of evolutionary interest stem from the human gene smim45. Web an open reading frame (orf) refers to a continuous dna (deoxyribonucleic acid) sequence with the potential to encode a protein. But they can also use orfs for more experimental purposes. [1] [2] translation of the uorf typically inhibits downstream expression of the primary orf.



![Open Reading Frame [ORF] Definition, Role in Protein Synthesis and](https://i2.wp.com/i.ytimg.com/vi/1T1S6NgECA0/maxresdefault.jpg)